Overview¶
A Module is a useful biochemical formulation, often incorporating a genetically encoded component, that performs a particular function. Modules Specifications answer the questions 1) “what is it?” and 2) “What should I expect when I implement it?”.
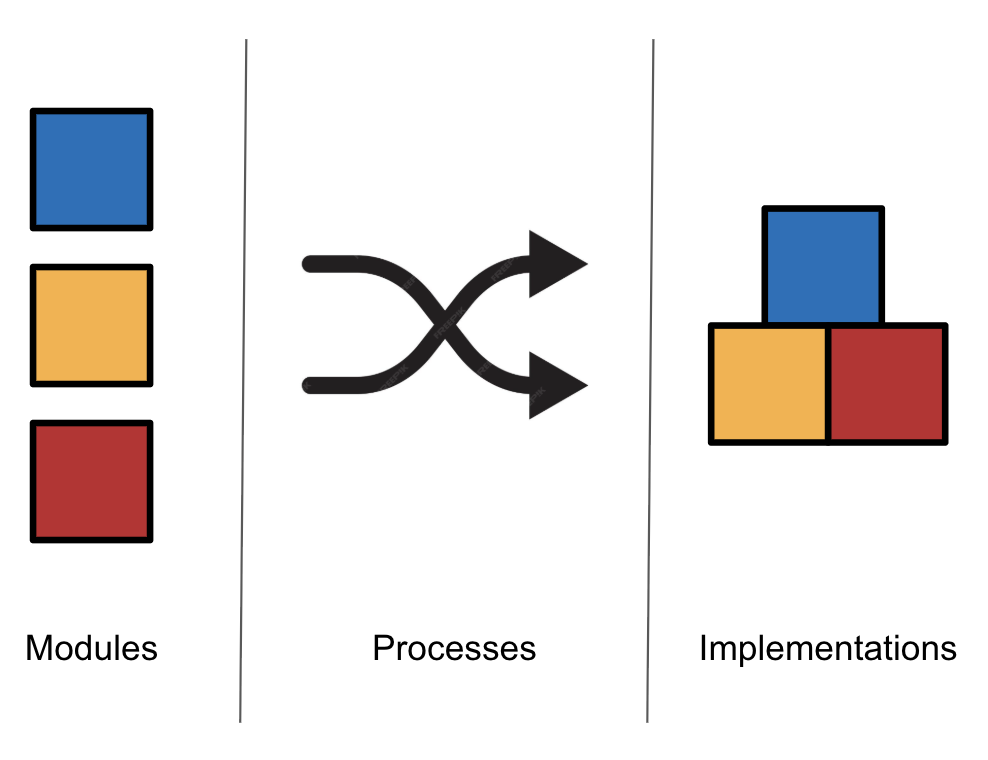
Modules can be combined with Process/Protocol to create Integrations.
Module Specifications contain the following information:
Brief description
Expected behavior
Where to access the materials
design file of genetic components, if applicable
schematic describing basic use
List of reference implementations
List of Modules¶
| Module Class | Module Implementation | Base Module | Status |
|---|---|---|---|
| Reporter | plamGFP | PURExpress Cell | Distribution |
| deGFP | Nucleus Cytosol; PURExpress Cell | Distribution | |
| Membrane | POPC/Chol | PURExpress Cell | Distribution |
| Membrane Pore | alpha-Hemolysin | PURExpress Cell | Distribution |
| Cx43 | PURExpress Cell | DevNote | |
| Detector | tetR-aTc | PURExpress Cell | Distribution |
| Emitter | IV-HSL | PURExpress Cell | Distribution |
| Energy | PPK | PURExpress Cell | DevNote |
| Control | ClpXP | PURExpress Cytosol | DevNote* |
| Chaperone | SecYEG | - | Planned |
Module contribution standards¶
Cytosol Module Standard