Overview¶
Presented here is our technical roadmap. These are modules that are either being actively worked on or planned for the near future.
Molecular sensing and response¶
An incredible power of synthetic cells is their ability to sense and respond to the environment with molecular precision. As engineers we would love to adapt, combine, and deploy these capabilities, but modifying natural systems is challenging—we don’t understand much of the biology and, even if we did, each system is unique.
Synthetic cells offer a defined context for developing sensors and responses into modular, composable systems, and can greatly expand what is possible to design and deploy with biology. For example, deploying sensors derived from multiple organisms to detect targets across a range of molecular types including small organics, RNAs, DNAs, proteins, sugars, and fats; producing amplified signals in response to detection of multiple targets via cytosolic signal processing and logic; or communicating with natural cells in the environment.
With Nucleus we are planning to port over the key modules associated with molecular sensing and response into Nucleus Cytosol and Cells.
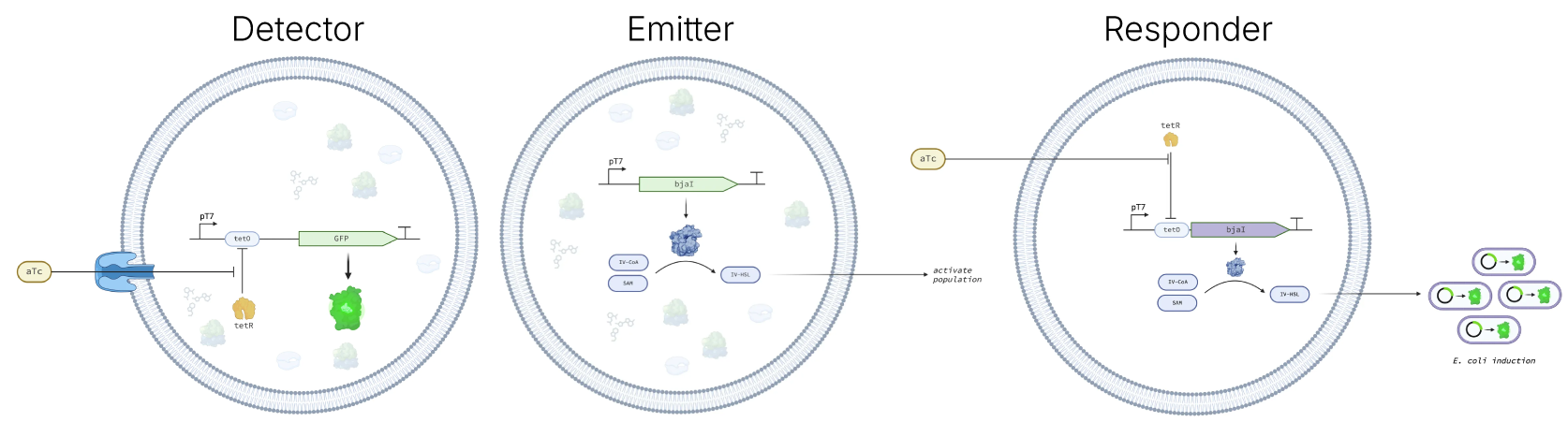
The Detector, Emitter, and Responder cells.
Developer cells¶
Advancing synthetic cell science and engineering to incorporate all essential functions of life and beyond, with fully understood components and emergent functions, will require a shared cell platform capable of integrating increasingly sophisticated cell modules from many developers. We have thus far introduced PURE-based synthetic cells as a baseline option for shared engineering, and demonstrated implementation of detector, emitter, and responder modules. While the PURE system forms an excellent, defined base cytosol for implementing simple modules in synthetic cells, its capacity for supporting sophisticated behaviors is quite limited. Critically, the system can only express proteins for a few hours until it runs out of energy and accumulates toxic waste products, with no ability to tune protein production or maintain energy and waste balance.
We are overcoming these challenges and enabling the shared development of increasingly capable synthetic cells by building a baseline “Developer Cell” that extends Nucleus PURE-based synthetic cells with integrated energy recycling, dynamic expression control, and membrane protein translation (Figure 1). These additional functions have individually already been demonstrated by the community. We will implement them as Nucleus modules and integrate them for use in the Nucleus Distribution. We will optimize the Developer Cell to support additional modules and produce tools and workflows to enable and accelerate further module integration and development. For more information, read out planning devnote Developer Cell: Project Introduction
With Nucleus we are planning to port over the energy and control modules into Nucleus Cytosols and Cells and begin planning the membrane protein translation module.
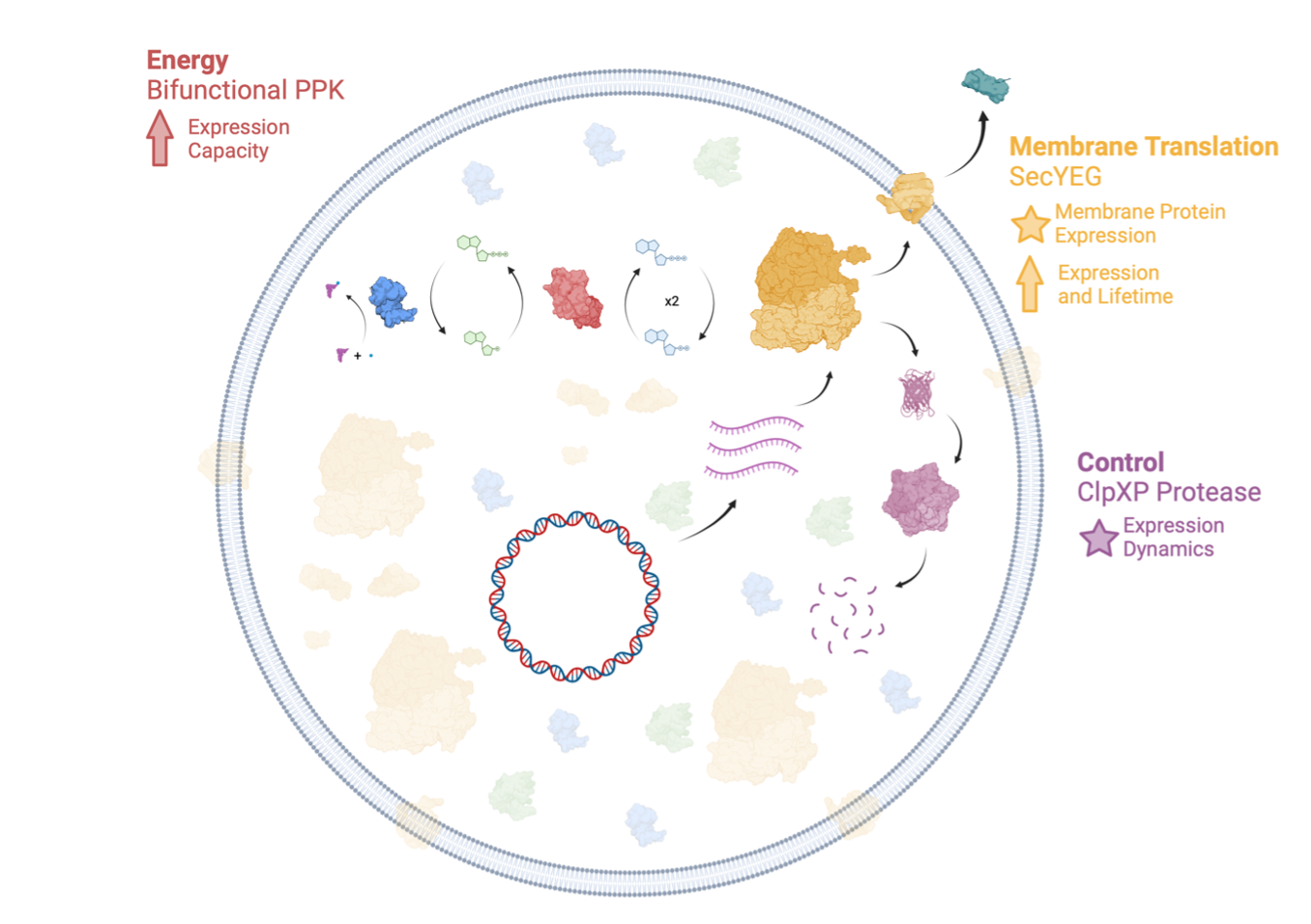
The Developer Cell. We are building an integrated chassis for synthetic cell engineering that incorporates energy generation, protein control, and membrane protein translation, needed for large-scale integration.
Long term vision¶
The long term goal for synthetic cells is to enable predictable engineering of complex “biomachines”, where a biomachine is an engineered system that makes use of biomolecules to carry out a useful function. See the SynCell Wiki for more information
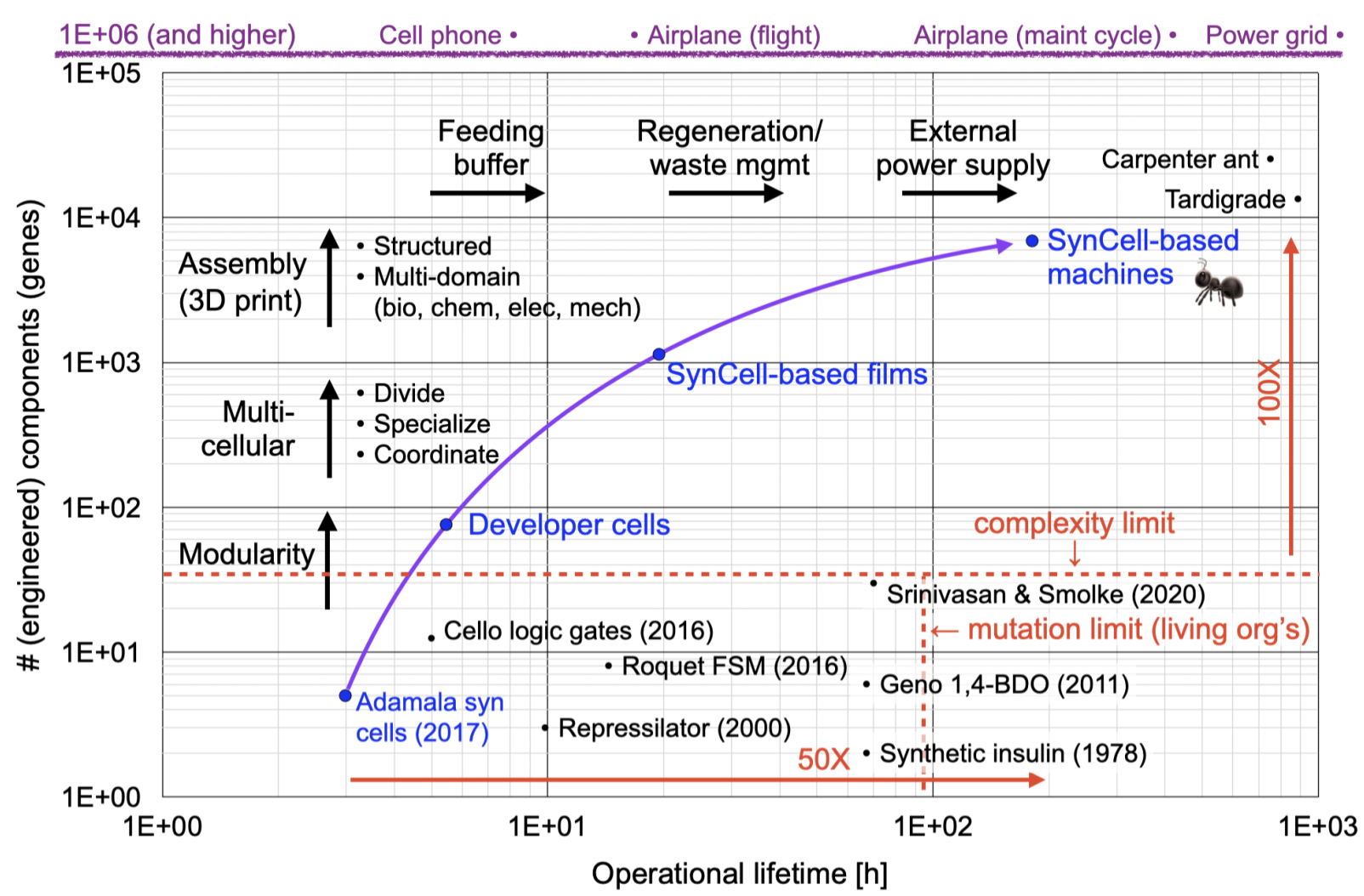
“White space” chart, showing a possible path to engineering biology at scale using synthetic cells. Figure by Richard Murray used under CC-BY-4.0 / unmodified from original.