This tutorial guides you through analyzing time-series fluorescence data from PURE (Protein synthesis Using Recombinant Elements) cell-free expression experiments. We’ll cover data loading, normalization, visualization, kinetic parameter fitting, and summary statistics.
1. Setup¶
First, import the necessary libraries and set up the plotting environment.
The custom platereader module that contains specialized functions for loading plate reader data, performing kinetic analysis, and visualization. The plot_setup() function configures matplotlib for time-series plots.
%load_ext autoreload
%autoreload 2
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# Import the cdk platereader module
from cdk.analysis.cytosol import platereader as pr
# Set up plotting
pr.plot_setup()2. Load Data¶
Load your plate reader data and merge it with the platemap that describes experimental conditions.
data_fileis the location of your platereader data (ideally in biotek-cdk format)platemap_file: location of your platemap CSV with Well and experimental conditionsload_platereader_data()reads your plate reader output file and parses it into a standardized format with the platemap integratedThe resulting DataFrame contains columns:
Well,Row,Column,Time,Seconds,Temperature (C),Read,Data, plus any columns from your platemapDatacolumn contains fluorescence measurements (RFU - Relative Fluorescence Units)Timerepresenting elapsed time
# Specify file paths
data_file = "sample-data/20251111-122213-cytation5-pure-timecourse-gfp-MFG-98-tRNA-QC-biotek-cdk.txt"
platemap_file = "sample-data/platemap.csv"
# Load data
data, platemap = pr.load_platereader_data(
data_file=data_file,
platemap_file=platemap_file,
platereader="biotek-cdk" # Options: "cytation", "envision", "biotek-cdk"
)
# Checkout first few rows
data.head()| Time | Well | Data | Row | Column | Read | Plate | Clock Time | Date | Experiment Name | ... | DNA ID | [DNA] (ng/uL) | PMix Vol (uL) | Ribosome Vol (uL) | SMS Vol (uL) | tRNA Vol (uL) | DNA Vol (uL) | RNase Inhib Vol (uL) | Water vol (uL) | Rxn Volume (uL) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 days 00:00:33 | B2 | 245 | B | 2 | GFP-Gext | Plate 1 | 2025-11-11 12:24:37 | 11/11/2025 | MFG-98-tRNA-QC | ... | AR-805 | 120.00 | 3.00 | NaN | 3.00 | 1.00 | 0.50 | 0.50 | 2.00 | 10.00 |
| 1 | 0 days 00:05:33 | B2 | 254 | B | 2 | GFP-Gext | Plate 1 | 2025-11-11 12:29:37 | 11/11/2025 | MFG-98-tRNA-QC | ... | AR-805 | 120.00 | 3.00 | NaN | 3.00 | 1.00 | 0.50 | 0.50 | 2.00 | 10.00 |
| 2 | 0 days 00:10:33 | B2 | 249 | B | 2 | GFP-Gext | Plate 1 | 2025-11-11 12:34:37 | 11/11/2025 | MFG-98-tRNA-QC | ... | AR-805 | 120.00 | 3.00 | NaN | 3.00 | 1.00 | 0.50 | 0.50 | 2.00 | 10.00 |
| 3 | 0 days 00:15:33 | B2 | 230 | B | 2 | GFP-Gext | Plate 1 | 2025-11-11 12:39:37 | 11/11/2025 | MFG-98-tRNA-QC | ... | AR-805 | 120.00 | 3.00 | NaN | 3.00 | 1.00 | 0.50 | 0.50 | 2.00 | 10.00 |
| 4 | 0 days 00:20:33 | B2 | 265 | B | 2 | GFP-Gext | Plate 1 | 2025-11-11 12:44:37 | 11/11/2025 | MFG-98-tRNA-QC | ... | AR-805 | 120.00 | 3.00 | NaN | 3.00 | 1.00 | 0.50 | 0.50 | 2.00 | 10.00 |
5 rows × 29 columns
4. Plot Raw Curves¶
Visualize the time-series fluorescence data to inspect curve shapes and identify any issues.
plot_curves_by_name()creates line plots with time on x-axis and fluorescence (Data) on y-axisEach line represents one well, colored by the
Namecolumn (experimental condition)If you have multiple gain data, they’ll be plotted in separate subplots (but you should choose one to move forward with)
This visualization helps you spot outliers, failed reactions, or unexpected kinetics before fitting
g = pr.plot_curves(data=data)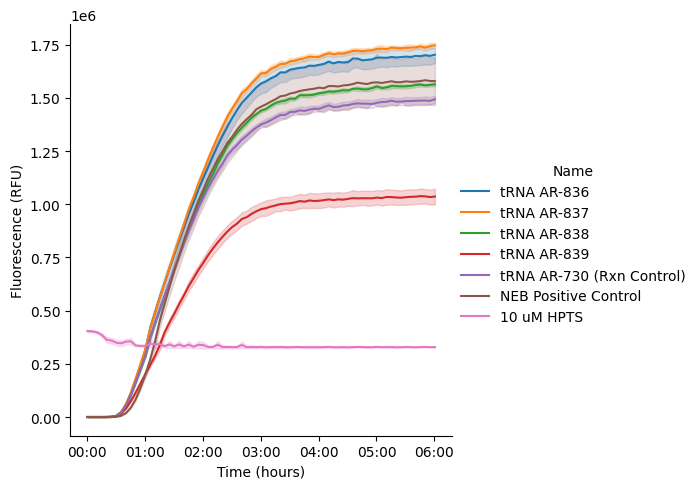
3. Normalize Data¶
Normalize your data to an internal fluorescence sample so that you have relative fluorescence to compare to other experiments.
data = pr.normalize_data_to_controls(data, ctrl_name = '10 uM HPTS')Data Normalized to 10 uM HPTS in col data_normalized. The active column for subsequent operations is: data_normalizedNow replot your curves to see them normalized
g = pr.plot_curves(data=data)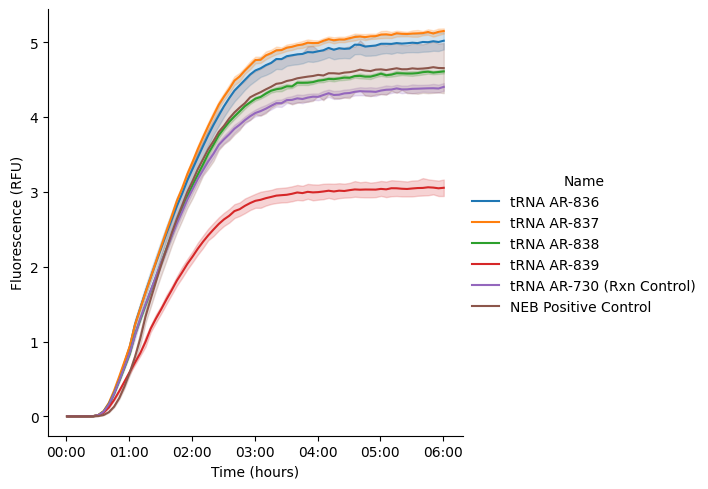
5. Kinetic Analysis¶
Fit sigmoid curves to the data to extract kinetic parameters: maximum velocity (Vmax), lag time, steady-state level, and drift.
kinetic_analysis()fits a sigmoid curve (with optional drift term) to each replicate wellThe model:
: Steady-state level (asymptote)
: Growth rate (steepness)
: Inflection point (time of max velocity)
: Drift rate (fluorescence change after steady-state)
: Drift onset time
Metrics extracted:
Vmax (
Velocity Max): Maximum rate of fluorescence increase (slope at inflection point)Lag time: Time to reach the exponential phase
Steady-state: Final fluorescence level and time to reach 95% of asymptote
Drift: Rate of signal decay or increase after steady-state
R²: Goodness of fit
# Perform kinetic analysis using sigmoid_drift model
kinetics = pr.kinetic_analysis(
data=data,
group_by=['Name'], # Group by experimental condition
)
kinetics.head()PROVIDING AVERAGED KINETICS| Velocity | Lag | Steady State | Fit | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Time | data_normalized | Max | Time | data_normalized | Time | data_normalized | params | R^2 | drift | |
| Name | ||||||||||
| NEB Positive Control | 0 days 01:34:45.616715696 | 2.25 | 2.69 | 0 days 00:44:28.688704778 | 0.30 | 0 days 02:48:47.196931504 | 4.28 | [4.501773951041968, 2.3866385500131293, 1.5793... | 1.00 | 0.08 |
| tRNA AR-730 (Rxn Control) | 0 days 01:29:01.540193308 | 2.19 | 2.34 | 0 days 00:32:45.112764155 | 0.19 | 0 days 02:51:52.382459217 | 4.17 | [4.384380953592719, 2.1324521783287356, 1.4837... | 1.00 | 0.07 |
| tRNA AR-836 | 0 days 01:31:43.799320428 | 2.52 | 2.56 | 0 days 00:32:30.333969522 | 0.21 | 0 days 02:58:55.280264960 | 4.79 | [5.0426505851597225, 2.0266336639016616, 1.528... | 1.00 | 0.07 |
| tRNA AR-837 | 0 days 01:32:27.273708334 | 2.58 | 2.66 | 0 days 00:34:09.146983246 | 0.24 | 0 days 02:58:17.284049550 | 4.90 | [5.161645765443068, 2.0583942112412883, 1.5409... | 1.00 | 0.07 |
| tRNA AR-838 | 0 days 01:31:47.921783514 | 2.31 | 2.41 | 0 days 00:34:09.156125597 | 0.22 | 0 days 02:56:39.983994240 | 4.39 | [4.622649794838434, 2.081776126751595, 1.52997... | 1.00 | 0.06 |
Visualize Fits on Individual Wells¶
What’s happening:
plot_kinetics_by_well()overlays fitted curves and kinetic parameters on raw dataVisual inspection ensures fits are reasonable (high R², smooth curves)
Annotations show where Vmax, lag time, and steady-state occur
# Plot kinetic fits
g, kinetics = pr.plot_kinetics(data, kinetics=kinetics, group_by=["Name"])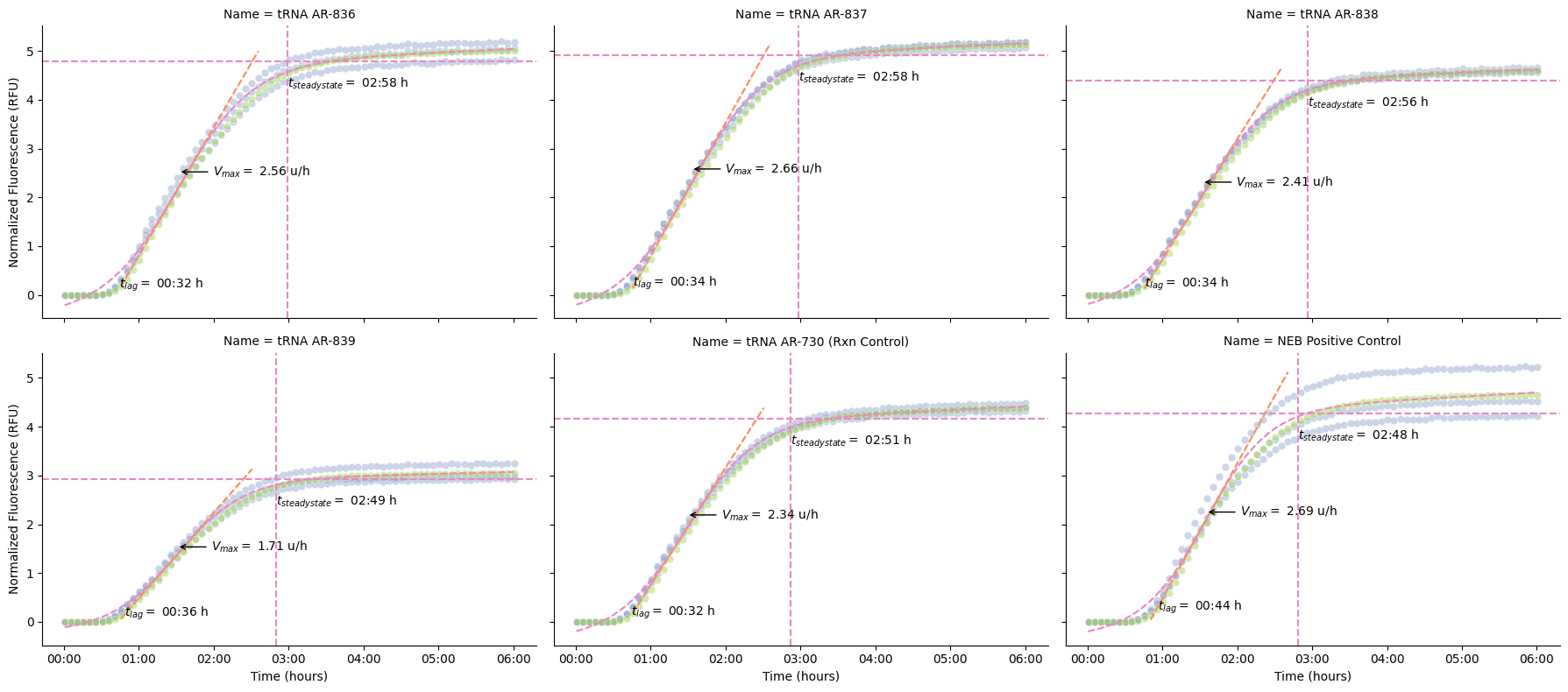
6. Summary Plots¶
Compare kinetic parameters across experimental conditions.
The
plot_kinetics_summary()function creates a comprehensive multi-panel figure with raw curves, steady-state, Vmax, and driftBar plots compare kinetic metrics across conditions
Steady-state: Final protein expression level (proxy for yield)
Vmax: Maximum rate of protein synthesis
Drift: Signal stability (photobleaching, aggregation, continued synthesis...etc)
Error bars show std across technical replicates
pr.plot_summary(data)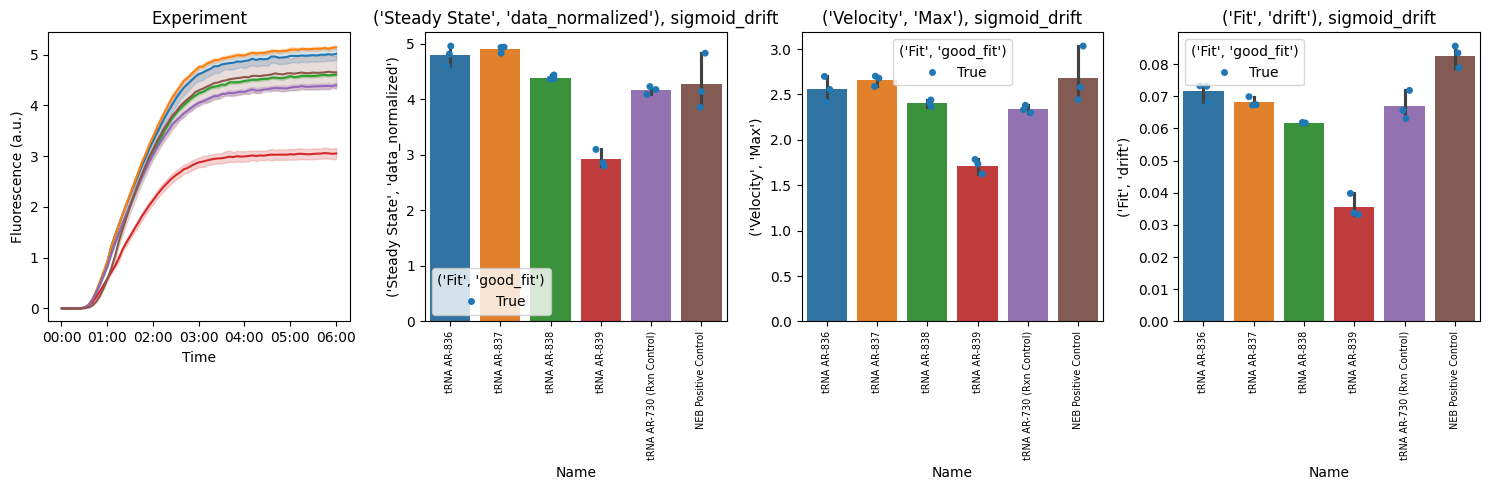
Key Metrics Explained¶
1. Steady-State Level (Steady State, Data)¶
The final fluorescence value reached by the reaction
Represents the total amount of protein produced
Higher values indicate greater expression yield
2. Maximum Velocity (Velocity, Max)¶
The steepest slope of the fluorescence curve (at the inflection point)
Units: RFU per second
Reflects the peak rate of protein synthesis
Sensitive to enzyme activity, substrate availability, and reaction conditions
3. Lag Time (Lag, Time)¶
Time before exponential fluorescence increase begins
May reflect time for ribosome assembly or initial translation steps
Shorter lag times suggest faster reaction initiation
4. Drift (Fit, drift)¶
Rate of fluorescence change after reaching steady-state
Positive drift: continued synthesis or aggregation
Negative drift: photobleaching, protein degradation, or quenching
Units: RFU per second
5. R² Value (Fit, R^2)¶
Goodness of fit (0 to 1, higher is better)
R² > 0.98 indicates excellent fit
Poor fits may indicate noisy data, overflow errors, or non-sigmoid kinetics
Tips and Troubleshooting¶
Overflow errors: Wells with
OVRFLWorNaNvalues are automatically excluded from fittingPoor fits (low R²): Inspect raw curves for anomalies (bubbles, evaporation, pipetting errors)
Drift: Sometimes seen in kinetics curves; use
sigmoid_driftmodelMultiple replicates: Always include technical replicates and report error bars
Comparing conditions: Normalize or blank data consistently across all samples
Next Steps¶
Export kinetics results:
pr.export_kinetics(kinetics, 'results.csv')Statistical analysis: Use
scipy.statsorstatsmodelsfor ANOVA/t-testsParameter optimization: Vary Mg²⁺, K⁺, or other conditions to maximize Vmax or steady-state
Mechanistic modeling: Fit ODE models to extract biological rate constants