Overview¶
The Nucleus Distribution allows you to implement an open standard for constructing synthetic cells based on Nucleus Cytosol, an open forumulation version of the PURE system.
The Distribution includes validated protocols, genetic constructs, software tools, datasets, and biological materials that are designed to work, and work together. All components of the Distribution are open source, allowing anyone to use and modify the components. As an open source project, the Distribution enabling the collaborative development of synthetic cells of increasing complexity.
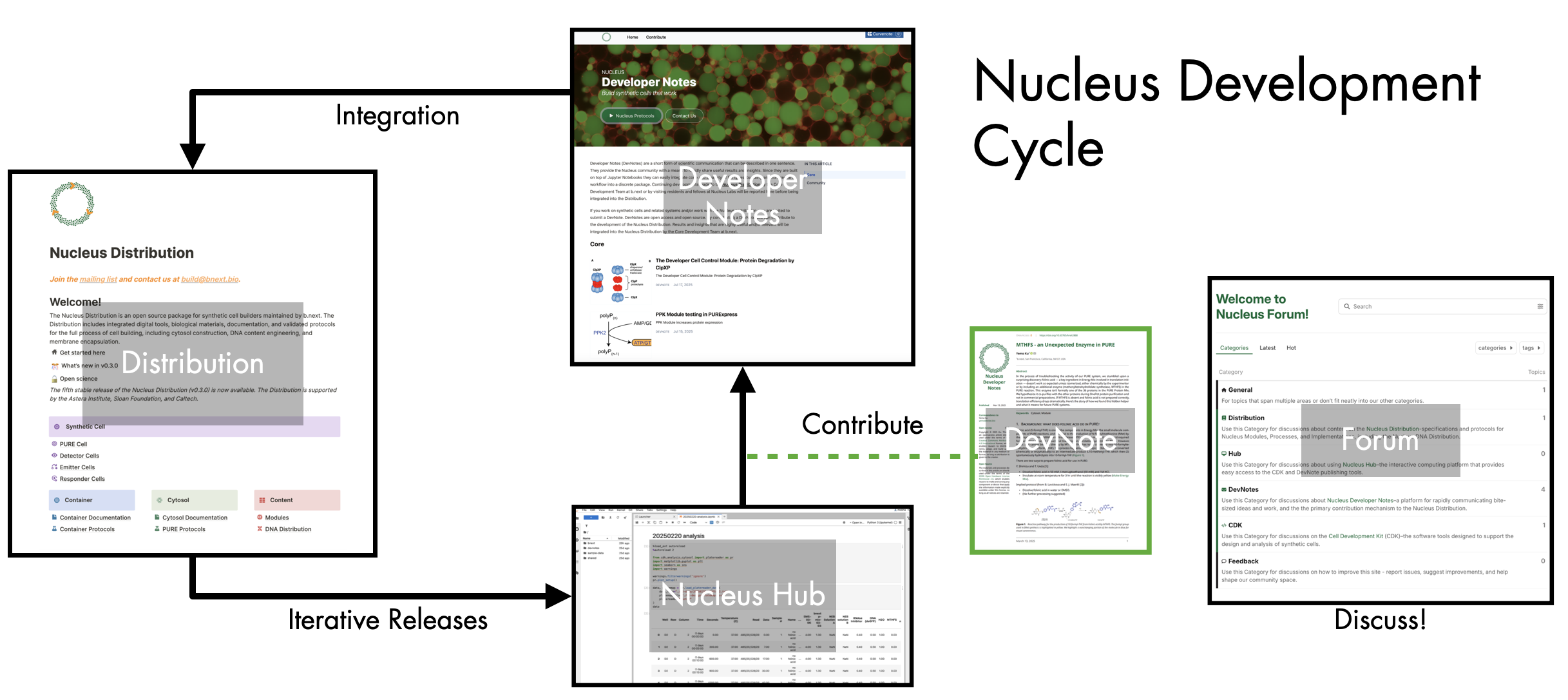
The Nucleus Development Cycle. All components of the Nucleus platform work together to support the collaborative development of increasingly reliable and sophisticated synthetic cells. The Distribution (this site) is a knowledge base that contains protocols and documentation for implementing validated synthetic cell modules. The Hub provides a resource for data sharing, analysis using standard digital tools. Useful results can be shared with the community as DevNotes. DevNotes can be viewed on the Developer Note website. Periodically, contributions made as Devnotes are integrated into the Distribution as validated protocols for others to use and build upon. All components of the platform can be discussed at the Forum, come say hello!
Structure of the Documentation¶
Get started¶
If you’re new to Nucleus, checkout From Zero to DevNote. That guide will walk you through the documentation.
Protocols¶
🧬 DNA Distribution
Physical materials that are used to implement Processes and Modules.
⚙️ Processes
Processes are core protocols for implementing base cytosol and cells.
🧱 Modules
Modules extend the functionality of base cytosol and cells.
🏗️ Implementations
Implementations are useful combinations of Modules and Processes.
Guides¶
TODO
Get in touch¶
Join the mailing list or contact us to request access to materials, get involved with the community, or ask questions.